Applications of CDI to Biology, Materials Science and Nanoscience
Biological Applications
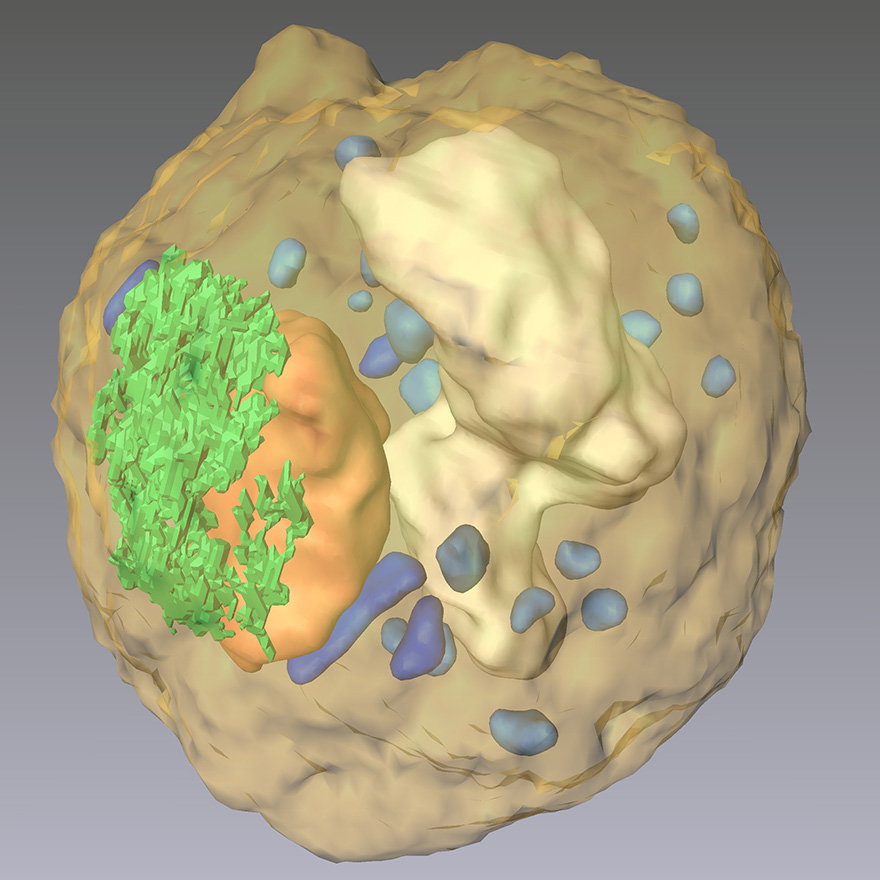 CDI reveals the 3D cellular organelles inside the yeast spore cell, showing nucleus (orange), ER (green), vacuole (white), mitochondria (blue), and granules (light blue). (Scale bar, 500 nm)
CDI reveals the 3D cellular organelles inside the yeast spore cell, showing nucleus (orange), ER (green), vacuole (white), mitochondria (blue), and granules (light blue). (Scale bar, 500 nm)
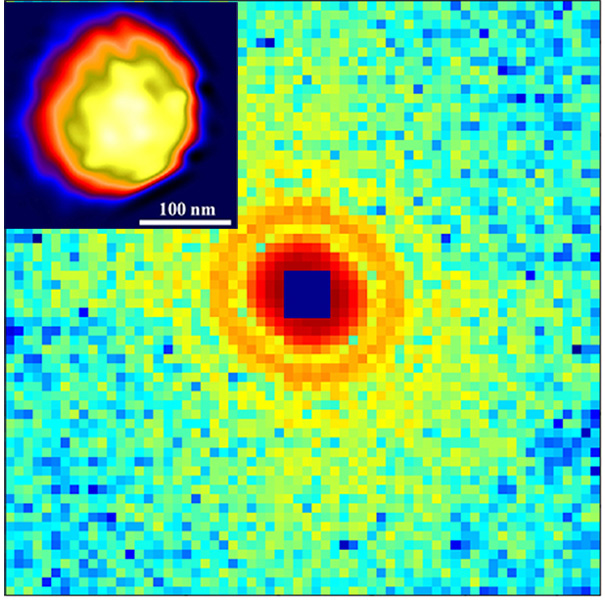 First coherent X-ray diffraction pattern measured from a single, unstained herpesvirus virion and its reconstructed structure (inset), where the capsid is in yellow.
First coherent X-ray diffraction pattern measured from a single, unstained herpesvirus virion and its reconstructed structure (inset), where the capsid is in yellow.
By using novel imaging technologies and labeling techniques, super-resolution fluorescence microscopy can study dynamic processes in living cells at the tens of nanometer level, but requires labeling of specific molecules, and can accommodate limited sample thickness. To achieve considerably higher resolution, electron microscopy is the method of choice, but is limited to imaging thinner samples owing to the short penetration depth of electrons. Compared with super-resolution and electron microscopy, CDI has three unique features. First, due to the large penetration depth of x-rays, CDI can image whole biological cells without the need of sectioning. Second, CDI takes advantage of the phase shift (contrast) of the intrinsic density in biological specimens that enables quantitative 3D imaging of the entire contents of cells and cellular organelles using their natural contrast. Finally, by avoiding the use of lenses, the resolution of CDI is limited only by how high a scattering angle (spatial frequency) the sample diffracts the illuminating light.
We for the first time applied CDI to image biological specimens in 2003 (1). Using coherent X-rays from a 3rd generation synchrotron radiation source, we measured diffraction patterns from intact E. coli bacteria and reconstructed their structures with a resolution of 30 nm. The distribution of proteins inside the bacteria labeled with manganese oxide has been identified and confirmed by fluorescence microscopy images (1). In 2010, we reported quantitative 3D diffractive imaging of a yeast spore cell at a resolution of 50–60 nm, and identified the 3D morphology and structure of cellular organelles including cell wall, vacuole, endoplasmic reticulum, mitochondria, granules, nucleus, and nucleolus inside the cell (2).In order to significantly improve the spatial resolution, cryogenic technology has to be implemented to keep the biological specimen at liquid nitrogen temperature. In 2015, we performed the first experimental demonstration of cryogenic CDI for quantitative 3D imaging of whole frozen-hydrated cells, which opens an important venue for revealing the 3D cellular architecture of whole cells in their natural state (3).
In addition to these important biological CDI experiments with synchrotron radiation, in 2001 we showed through numerical simulations that the diffraction patterns accumulated from multiple copies of single rubisco biomolecules, each generated by a femtosecond XFEL pulse, can be successfully phased and transformed into an accurate electron density map with 2.5-Å resolution (4). This work became a scientific justification for the construction of the Linac Coherent Light Source at SLAC National Accelerator Laboratory and other XFELs around the world. To experimentally demonstrate the feasibility of imaging single large protein complexes, we performed the first diffractive imaging of single, unstained viruses using coherent X-rays (5). Many groups around the world have now applied CDI to image a wide range of biological specimens such as large protein complexes, viruses, cellular organelles, and whole cells using synchrotron radiation, XFELs and HHG sources (6-8).
Materials Science and Nanoscience Applications
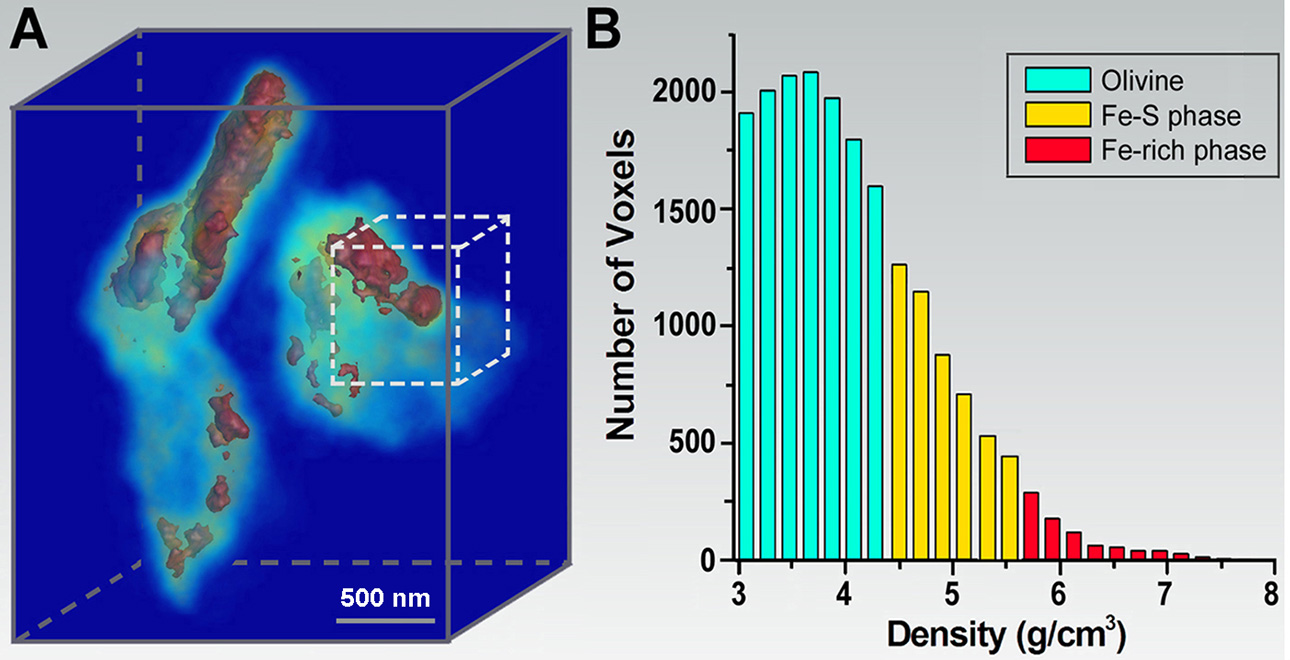 (A) 3D morphology of Fe-rich and Fe-S phases in an olivine matrix. (B) Histogram of the Fe-rich phase, Fe-S phase, and olivine distribution within a dotted-line cube in (A).
(A) 3D morphology of Fe-rich and Fe-S phases in an olivine matrix. (B) Histogram of the Fe-rich phase, Fe-S phase, and olivine distribution within a dotted-line cube in (A).
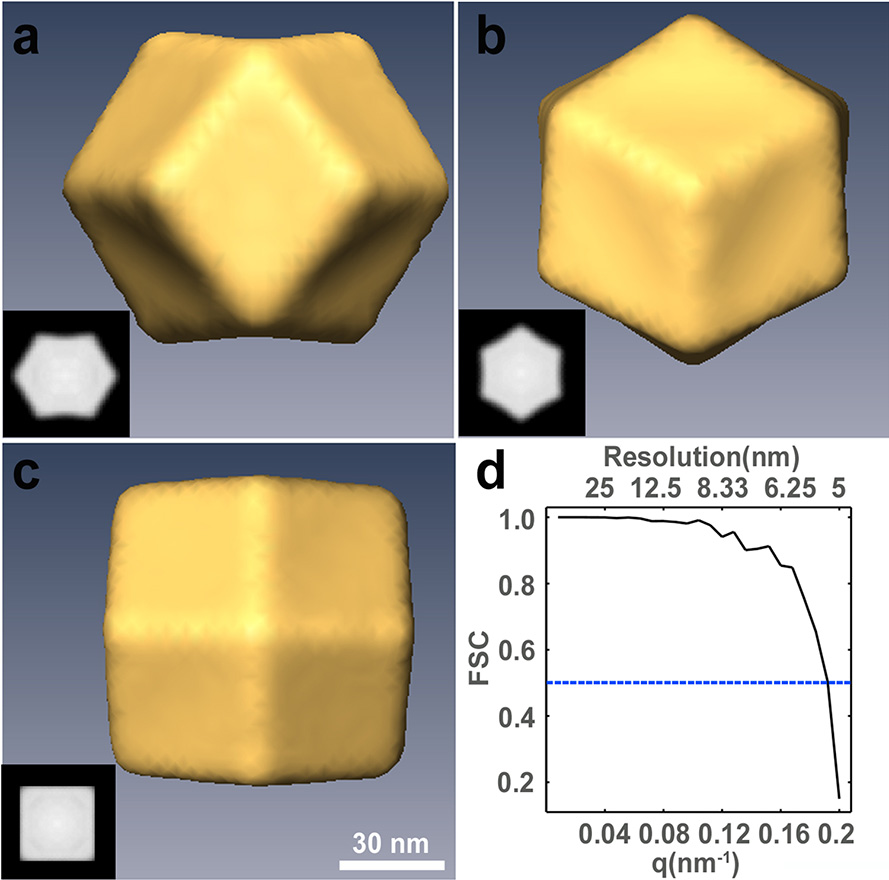 Single-shot 3D structure determination. (a-c) Iso-surface renderings of the final 3D reconstruction of a trisoctahedral Au nanocrystals along the 2-, 3- and 4-fold rotational symmetry, respectively. The insets show 3.3-nm-thick centro-slices of the 3D reconstruction along the corresponding directions. (d) Fourier shell correlation (FSC) comparison between two independently reconstructed gold nanocrystals. Based on the FSC = 0.5 criterion, a 3D resolution of the reconstructions was estimated to be ~5.5 nm.
Single-shot 3D structure determination. (a-c) Iso-surface renderings of the final 3D reconstruction of a trisoctahedral Au nanocrystals along the 2-, 3- and 4-fold rotational symmetry, respectively. The insets show 3.3-nm-thick centro-slices of the 3D reconstruction along the corresponding directions. (d) Fourier shell correlation (FSC) comparison between two independently reconstructed gold nanocrystals. Based on the FSC = 0.5 criterion, a 3D resolution of the reconstructions was estimated to be ~5.5 nm.
Because X-rays have a larger penetration depth than electrons and destructive sample preparation can often be avoided, CDI is ideally suited for quantitative 3D characterization of materials at the nanoscale. In 2002, we carried out the first 3D CDI experiment – imaging the 3D structure of a non-crystalline material at 50 nm resolution (9). Subsequently, we performed quantitative 3D imaging of a heat-treated GaN particle with a voxel size of 17x17x17 nm3. The platelet structure of GaN and the formation of small islands on the surface of the platelets were observed and the internal GaN-Ga2O3 core shell structure was imaged in 3D (10). In 2008, we applied CDI to image mineral crystals inside biological composite materials - intramuscular fish bone - at the nanometer scale resolution. Mineral crystals in collagen fibrils were identified at different stages of mineralization. Based on the experimental results and biomineralization analyses, a dynamic model was proposed to account for the nucleation and growth of mineral crystals in the collagen matrix (11). More recently, we performed quantitative diffractive imaging of a molten Fe-rich alloy and crystalline olivine sample, synthesized at 6 GPa and 1800 °C, with a 3D resolution of 30-50 nm (12). The 3D mass density map was determined and the 3D distribution of the Fe-rich and Fe-S phases in the olivine-Fe-S sample was observed. These results indicate that the Fe-rich melt exhibits varied 3D shapes and sizes in the olivine matrix. This sequence of experiments paves the way for quantitative 3D characterization of materials at nanoscale resolution under extreme pressures and temperatures.
In 2010, we also developed a new imaging approach, termed ankylography (derived from the Greek words ankylos meaning ‘curved’ and graphein meaning ‘writing’), which under certain circumstances enables complete 3D structure determination from a single exposure (13). An important implication of this work is its ability to achieving single-shot 3D structure determination with XFEL pulses. In 2014, we reported the experimental demonstration of single-shot 3D structure determination of individual gold nanocrystals at 5.5 nm resolution using 10 fs XFEL pulses (14). Coherent diffraction patterns were collected from high-index-faceted nanocrystals, each struck by an XFEL pulse. Taking advantage of the symmetry of the nanocrystal and the curvature of the Ewald sphere, the 3D structure of each nanocrystal was reconstructed from a single-shot diffraction pattern. The significance of this work is twofold. First, as symmetry exists in many nanocrystals and virus particles, this single-shot 3D imaging method can be applied to 3D structure studies of such particles at the nanometer resolution on femtosecond time scales. Second, with enough copies of identical nanocrystals, the combination of symmetry, the curvature of the Ewald sphere and CDI can potentially be used to determine the 3D structure of nanocrystals at atomic resolution.
Selected Publications
1. J. Miao, K. O. Hodgson, T. Ishikawa, C. A. Larabell, M. A. LeGros and Y. Nishino, "Imaging Whole Escherichia Coli Bacteria by Using Single Particle X-ray Diffraction", Proc. Natl. Acad. Sci. USA 100, 110-112 (2003).
2. H. Jiang, C. Song, C.-C. Chen, R. Xu, R., K. S. Raines, B. P. Fahimian, C. Lu,. T.-H. Lee, A. Nakashima, J. Urano, T. Ishikawa, F. Tamanoi, J. Miao, "Quantitative 3D Imaging of Whole, Unstained Cells by Using X-ray Diffraction Microscopy", Proc. Natl. Acad. Sci. USA 107, 11234–11239 (2010).
3. J. A. Rodriguez, R. Xu, C.-C. Chen, Z. Huang, H. Jiang, A. L. Chen, K. S. Raines, A. Pryor, Jr., D, Nam, L. Wiegart, C. Song, A. Madsen, Y. Chushkin, F. Zontone, P. J. Bradley and J. Miao, "Three-dimensional Coherent X-ray Diffractive Imaging of Whole, Frozen-Hydrated Cells", IUCrJ 2, 575–583 (2015).
4. J. Miao, K. O. Hodgson and D. Sayre, "An approach to three-dimensional structures of biomolecules by using single-molecule diffraction images", Proc. Natl. Acad. Sci. USA 98, 6641-6645 (2001).
5. C. Song, H. Jiang, A. Mancuso, B. Amirbekian, L. Peng, R. Sun, S. S Shah, Z. H. Zhou, T. Ishikawa and J. Miao, "Quantitative Imaging of Single, Unstained Viruses with Coherent X-rays", Phys. Rev. Lett. 101, 158101 (2008).
6. J. Miao, T. Ishikawa, I. K. Robinson and M. M. Murnane, "Beyond crystallography: Diffractive imaging using coherent x-ray light sources", Science 348, 530-535 (2015).
7. M. Gallagher-Jones, J. A. Rodriguez and J. Miao, "Frontier Methods in Coherent X-ray Diffraction for High-Resolution Structure Determination", Q. Rev. Biophys. 49, e20 (2016).
8. I. Schlichting and J. Miao, "Emerging opportunities in structural biology with X-ray free-electron lasers", Curr. Opin. Struct. Biol. 22, 613–626 (2012).
9. J. Miao, T. Ishikawa, B. Johnson, E. H. Anderson, B. Lai and K. O. Hodgson, "High Resolution 3D X-ray Diffraction Microscopy", Phys. Rev. Lett. 89, 088303 (2002).
10. J. Miao, C. C. Chen, C. Song, Y. Nishino, Y. Kohmura, T. Ishikawa, D. Ramunno-Johnson, T. K. Lee and S. H. Risbud, "Three-Dimensional GaN-Ga2O3 Core Shell Structure Revealed by X-ray Diffraction Microscopy", Phys. Rev. Lett. 97, 215503 (2006).
11. H. Jiang, D. Ramunno-Johnson, C. Song, B. Amirbekian, Y. Kohmura, Y. Nishino, Y. Takahashi, T. Ishikawa and J. Miao, "Nanoscale Imaging of Mineral Crystals inside Biological Composite Materials Using X-ray Diffraction Microscopy", Phys. Rev. Lett. 100, 038103 (2008).
12. H. Jiang, R. Xu, C.-C. Chen, W. Yang, J. Fan, X. Tao, C. Song, Y. Kohmura, T. Xiao, Y. Wang, Y. Fei, T. Ishikawa, W. L. Mao and J. Miao, "Three-Dimensional Coherent X-Ray Diffraction Imaging of Molten Iron in Mantle Olivine at Nanoscale Resolution", Phys. Rev. Lett. 110, 205501 (2013).
13. K. S. Raines, S. Salha, R. L. Sandberg, H. Jiang, J. A. Rodríguez, B. P. Fahimian, H. C. Kapteyn, J. Du and J. Miao, "Three-dimensional structure determination from a single view", Nature 463, 214-217 (2010).
14. R. Xu, H. Jiang, C. Song, J. A. Rodriguez, Z. Huang, C.-C. Chen, D. Nam, J. Park, M. Gallagher-Jones, S. Kim, S. Kim, A. Suzuki, Y. Takayama, T. Oroguchi, Y. Takahashi, J. Fan, Y. Zou, T. Hatsui, Y. Inubushi, T. Kameshima, K. Yonekura, K. Tono, T. Togashi, T. Sato, M. Yamamoto, M. Nakasako, M. Yabashi, T. Ishikawa and J. Miao, "Single-shot three-dimensional structure determination of nanocrystals with femtosecond X-ray free-electron laser pulses", Nature Commun. 5, 4061 (2014).